Difference between revisions of "Contrib:Projectionist/Extra01"
(→Doing the study) |
m (Reverted edit of Atywehacyg, changed back to last version by Newacct) |
||
| (16 intermediate revisions by 3 users not shown) | |||
| Line 6: | Line 6: | ||
=== Preface === | === Preface === | ||
| − | If there is the need to analyze lots of topological likewise geometries then automation | + | If there is the need to analyze lots of topological likewise geometries then automation can make life easier. This paragraph describes automatable analysis of a 2D part section. Given the case the geometry comes as list of vertices in a known order we can read this list with a script and create a TUI script. |
=== The input data === | === The input data === | ||
| Line 34: | Line 34: | ||
| + | === Running the TUI script === | ||
| + | |||
| + | Salome will be started in terminal mode and the TUI script will be directly imported by command line option. As this whole analysis is made by one python script, only the parts concerning Salome will be shown here. The whole script will be listed somewhere below. | ||
| + | |||
| + | p1 = Popen("source /home/gerhard/salome_5.1.2/KERNEL_5.1.2/salome.sh", shell=True) | ||
| + | sts1 = os.waitpid(p1.pid, 0) | ||
| + | p2 = Popen("/home/gerhard/salome_5.1.2/KERNEL_5.1.2/bin/salome/runSalome -t -u ./outTUI.py", shell=True) | ||
| + | sts2 = os.waitpid(p2.pid, 0) | ||
| + | |||
| + | '''Popen()''' executes the given string as a sub-process. '''waitpid()''' ensures the sub-process is finished before the calling process continues execution. | ||
| + | |||
| + | === Creating the FE study === | ||
| + | |||
| + | This part was taken from CAELinux. It is the core of the CreateJob.py script that uses a GUI for the major input. Here the original script is shown in action. | ||
| + | |||
| + | [[Image:View_asterHelpers_createFeStudy_detail.png]] | ||
| + | |||
| + | I removed the things related to the GUI. Crude work I have to admit. This script receives the filename of the data file as argument. | ||
| + | |||
| + | |||
| + | #!/usr/bin/env python | ||
| + | # Create a new FE analysis job from a template | ||
| + | # J.Cugnoni, CAELinux.com, 2005 | ||
| + | # edited by G. Holzinger, JKU Linz, 2009 | ||
| + | |||
| + | import os | ||
| + | import os.path | ||
| + | import sys | ||
| + | |||
| + | fileName = sys.argv[1] | ||
| + | |||
| + | name = fileName[:fileName.rfind(".")] | ||
| + | |||
| + | templateASTK=""" | ||
| + | etude,fich,3,FR F | ||
| + | opt_val,rep_dex _VIDE | ||
| + | etude,fich,3,UL 8 | ||
| + | etude,fich,2,type mess | ||
| + | etude,fich,4,donnee 0 | ||
| + | option,nbmaxnook 1 | ||
| + | etude,fich,2,resultat 1 | ||
| + | nom_fich_export _VIDE | ||
| + | option,rep_dex 0 | ||
| + | etude,fich,4,compress 0 | ||
| + | etude oui | ||
| + | debug 0 | ||
| + | opt_val,cpresok RESNOOK | ||
| + | forlib_delete non | ||
| + | etude,fich,4,serv Local | ||
| + | etude,fich,3,donnee 0 | ||
| + | serv_fich_export -1 | ||
| + | surcharge,nbfic 0 | ||
| + | etude,fich,6,FR F | ||
| + | path_etude [__prjdir__] | ||
| + | option,rep_outils 0 | ||
| + | etude,fich,6,UL 80 | ||
| + | etude,fich,3,resultat 1 | ||
| + | option,cpresok 1 | ||
| + | etude,fich,5,compress 0 | ||
| + | etude,fich,1,FR F | ||
| + | etude,fich,1,serv Local | ||
| + | consult_supprimer non | ||
| + | memoire 128 | ||
| + | etude,fich,1,UL 20 | ||
| + | onglet_actif etude | ||
| + | asquit non | ||
| + | etude,fich,2,donnee 0 | ||
| + | option,classe 1 | ||
| + | etude,fich,4,type erre | ||
| + | consult_a_corriger non | ||
| + | ident non | ||
| + | etude,fich,4,resultat 1 | ||
| + | etude,fich,6,compress 0 | ||
| + | etude,fich,1,donnee 1 | ||
| + | rex non | ||
| + | etude,fich,1,type libr | ||
| + | etude,fich,1,nom ./[__prjname__]mesh.med | ||
| + | suivi_interactif 1 | ||
| + | path_sources _VIDE | ||
| + | pre_eda non | ||
| + | etude,fich,4,FR F | ||
| + | forlib_create non | ||
| + | etude,fich,4,UL 9 | ||
| + | etude,fich,3,nom ./[__prjname__].resu | ||
| + | serv_tests -1 | ||
| + | etude,fich,6,serv Local | ||
| + | etude,fich,5,nom ./[__prjname__].base | ||
| + | make_etude run | ||
| + | option,depart 1 | ||
| + | nom_profil [__astkfile__] | ||
| + | args _VIDE | ||
| + | etude,fich,5,resultat 1 | ||
| + | etude,fich,0,donnee 1 | ||
| + | opt_val,mem_aster _VIDE | ||
| + | etude,fich,3,serv Local | ||
| + | etude,fich,0,compress 0 | ||
| + | option,mem_aster 1 | ||
| + | asdeno non | ||
| + | serv_surcharge -1 | ||
| + | serv_sources -1 | ||
| + | surcharge non | ||
| + | etude,fich,6,type rmed | ||
| + | etude,fich,0,serv Local | ||
| + | M_1 oui | ||
| + | serv_etude -1 | ||
| + | M_2 non | ||
| + | etude,fich,6,resultat 1 | ||
| + | etude,fich,2,FR F | ||
| + | opt_val,rep_outils _VIDE | ||
| + | M_3 non | ||
| + | etude,fich,2,UL 6 | ||
| + | path_surcharge _VIDE | ||
| + | consult non | ||
| + | M_4 non | ||
| + | asno non | ||
| + | etude,fich,3,type resu | ||
| + | etude,fich,1,compress 0 | ||
| + | opt_val,ncpus 1 | ||
| + | emis_sans non | ||
| + | serveur localhost | ||
| + | opt_val,classe _VIDE | ||
| + | opt_val,dbgjeveux _VIDE | ||
| + | option,ncpus 1 | ||
| + | etude,fich,0,type comm | ||
| + | opt_val,facmtps 1 | ||
| + | etude,fich,0,resultat 0 | ||
| + | opt_val,rep_mat _VIDE | ||
| + | special _VIDE | ||
| + | tests,nbfic 0 | ||
| + | temps 240 | ||
| + | option,dbgjeveux 0 | ||
| + | etude,fich,5,FR R | ||
| + | etude,fich,5,serv Local | ||
| + | asrest non | ||
| + | batch 0 | ||
| + | etude,fich,5,UL 0 | ||
| + | etude,fich,2,compress 0 | ||
| + | option,facmtps 1 | ||
| + | serv_profil -1 | ||
| + | etude,fich,6,donnee 0 | ||
| + | etude,fich,0,FR F | ||
| + | option,rep_mat 0 | ||
| + | etude,fich,0,UL 1 | ||
| + | etude,fich,2,serv Local | ||
| + | etude,fich,0,nom ./[__prjname__].comm | ||
| + | asverif non | ||
| + | etude,fich,2,nom ./[__prjname__].mess | ||
| + | opt_val,depart _VIDE | ||
| + | etude,fich,1,resultat 0 | ||
| + | sources,nbfic 0 | ||
| + | etude,fich,5,donnee 0 | ||
| + | etude,fich,4,nom ./[__prjname__].erre | ||
| + | tests non | ||
| + | noeud localhost | ||
| + | etude,fich,5,type base | ||
| + | etude,fich,3,compress 0 | ||
| + | emis_prof non | ||
| + | etude,fich,6,nom ./[__prjname__]res.med | ||
| + | etude,nbfic 7 | ||
| + | agla non | ||
| + | opt_val,nbmaxnook 5 | ||
| + | version STA8.1 | ||
| + | path_tests _VIDE | ||
| + | nom_profil %s | ||
| + | path_etude %s | ||
| + | etude,fich,0,nom ./%s.comm | ||
| + | etude,fich,1,nom ./%smesh.med | ||
| + | etude,fich,2,nom ./%s.mess | ||
| + | etude,fich,3,nom ./%s.resu | ||
| + | etude,fich,4,nom ./%s.erre | ||
| + | etude,fich,5,nom ./%s.base | ||
| + | etude,fich,6,nom ./%sres.med | ||
| + | """ | ||
| + | |||
| + | def validate(): | ||
| + | bdir=vars["basedirname"].strip() | ||
| + | prjname=vars["prjname"].strip() | ||
| + | meshname=vars["meshname"].strip() | ||
| + | tplname=vars["tplname"].strip() | ||
| + | prjdir=os.path.join(bdir,prjname) | ||
| + | if os.path.exists(prjdir): | ||
| + | print "project name already exists in base directory" | ||
| + | else: | ||
| + | # project directory | ||
| + | os.makedirs(prjdir) | ||
| + | # file names | ||
| + | commfile=os.path.join(prjdir,prjname + ".comm") | ||
| + | messfile=os.path.join(prjdir,prjname + ".mess") | ||
| + | errefile=os.path.join(prjdir,prjname + ".erre") | ||
| + | resufile=os.path.join(prjdir,prjname + ".resu") | ||
| + | mmedfile=os.path.join(prjdir,prjname + "mesh.med") | ||
| + | rmedfile=os.path.join(prjdir,prjname + "res.med") | ||
| + | basefile=os.path.join(prjdir,prjname + ".base") | ||
| + | astkfile=os.path.join(prjdir,prjname + ".astk") | ||
| + | # copy files | ||
| + | from shutil import copyfile | ||
| + | copyfile(meshname,mmedfile) | ||
| + | copyfile(tplname,commfile) | ||
| + | # create ASTK profile | ||
| + | fd=open(astkfile,"w") | ||
| + | fd.write(templateASTK % ((astkfile,prjdir,) + (prjname,)*7)) | ||
| + | fd.close() | ||
| + | # show message and exit | ||
| + | print "New FE analysis project created successfully." | ||
| + | |||
| + | return 0 | ||
| + | |||
| + | vars={"basedirname": os.getcwd() + "/", | ||
| + | "prjname": name, | ||
| + | "meshname": "mesh_" + name + ".med", | ||
| + | "tplname": "command.comm"} | ||
| + | |||
| + | validate() | ||
| + | |||
| + | === Create the *.export file === | ||
| + | |||
| + | For this task there is also a script. This script receives the name of the study as argument. To adjust this file for your needs compare the output of this script with a *.export file from a running CodeAster study created by ASTK. Adjust everything you need to obtain equal *.export files. | ||
| + | |||
| + | |||
| + | #!/usr/bin/env python | ||
| + | # | ||
| + | # Create an .export-file for CodeAster | ||
| + | # G. Holzinger, JKU Linz, 2009 | ||
| + | |||
| + | import sys | ||
| + | import os | ||
| + | |||
| + | user = os.getlogin() | ||
| + | machine = os.uname()[1] | ||
| + | workingRoot = os.getcwd() | ||
| + | |||
| + | # read argument | ||
| + | name = sys.argv[1] | ||
| + | |||
| + | # open the file | ||
| + | f = open("./" + name + "/" + name + ".export",'w') | ||
| + | |||
| + | f.write("P profastk " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + ".astk\n") | ||
| + | f.write("P serveur localhost\n") | ||
| + | f.write("P noeud " + machine + "\n") | ||
| + | f.write("P username " + user + "\n") | ||
| + | f.write("P mclient " + machine + "\n") | ||
| + | f.write("P uclient " + user + "\n") | ||
| + | f.write("P version STA9.4\n") | ||
| + | f.write("P lang en\n") | ||
| + | f.write("P debug nodebug\n") | ||
| + | f.write("P mode interactif\n") | ||
| + | f.write("P ncpus 1\n") | ||
| + | f.write("P mpi_nbcpu 1\n") | ||
| + | f.write("P mpi_nbnoeud 1\n") | ||
| + | f.write("P classe \n") | ||
| + | f.write("P depart \n") | ||
| + | f.write("P distrib \n") | ||
| + | f.write("P timeout \n") | ||
| + | f.write("P exectool \n") | ||
| + | f.write("P nomjob " + name + "\n") | ||
| + | f.write("P origine ASTK 1.7.1\n") | ||
| + | f.write("P display " + machine + ":0.0\n") | ||
| + | f.write("A args \n") | ||
| + | f.write("A memjeveux 32.0\n") | ||
| + | f.write("P mem_aster 100.0\n") | ||
| + | f.write("A tpmax 240\n") | ||
| + | f.write("P memjob 131072\n") | ||
| + | f.write("P tpsjob 4\n") | ||
| + | f.write("P nbmaxnook 5\n") | ||
| + | f.write("P cpresok RESNOOK\n") | ||
| + | f.write("P facmtps 1\n") | ||
| + | f.write("P corefilesize unlimited\n") | ||
| + | f.write("F comm " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + ".comm D 1\n") | ||
| + | f.write("F libr " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + "mesh.med D 20\n") | ||
| + | f.write("F mess " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + ".mess R 6\n") | ||
| + | f.write("F resu " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + ".resu R 8\n") | ||
| + | f.write("F erre " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + ".erre R 9\n") | ||
| + | f.write("R base " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + ".base R 0\n") | ||
| + | f.write("F rmed " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + "res.med R 80\n") | ||
| + | f.write("P consbtc oui\n") | ||
| + | f.write("P soumbtc oui\n") | ||
| + | f.write("P actions make_etude") | ||
| + | |||
| + | # close the file | ||
| + | f.close() | ||
| + | |||
| + | === Run the FE study with Code Aster === | ||
| + | |||
| + | Here are only the few lines related to CodeAser shown, the whole script is listed further below. | ||
| + | |||
| + | exportFullPath = os.getcwd() + ("/" + name)*2 + ".export" | ||
| + | |||
| + | p5 = Popen("/opt/SALOME-MECA-2009.1-GPL/aster/outils/as_run --run " + exportFullPath, shell=True) | ||
| + | sts5 = os.waitpid(p5.pid, 0) | ||
=== Doing the study === | === Doing the study === | ||
| − | Here is where all the magic happens. | + | Here is where all the magic happens. Don't get confused with all the boxes, this source code belongs to one file. However these boxes subdividing the code into blocks may improve readability. |
| Line 102: | Line 392: | ||
# create mesh | # create mesh | ||
| − | p1 = Popen("source /SALOME-ROOT/salome_5.1.2/KERNEL_5.1.2/salome.sh", shell=True) | + | p1 = Popen("source /SALOME-ROOT/salome_5.1.2/KERNEL_5.1.2/salome.sh; /SALOME-ROOT/salome_5.1.2/KERNEL_5.1.2/bin/salome/runSalome -t -u ./outTUI.py", shell=True) |
sts1 = os.waitpid(p1.pid, 0) | sts1 = os.waitpid(p1.pid, 0) | ||
| − | |||
| − | |||
| − | if | + | if sts1[1] == 0: |
# create finite element study | # create finite element study | ||
| Line 180: | Line 468: | ||
usage() | usage() | ||
| + | |||
| + | The first Block is made up of comments. The most important thing here is the first line. It tells the linux shell what to do with this script. | ||
| + | |||
| + | In the second Block several python modules are imported. | ||
| + | |||
| + | The third block defines a functions that prints the usage information. Rudimentay though. | ||
| + | |||
| + | In the fourth block the number of arguments is determined. | ||
| + | |||
| + | The next three Blocks are dedicated to three possible cases. No arguments were passed, one - which is the correct number - or too many. | ||
| + | If the wrong number of arguments passed is wrong, then error messages will be printed. | ||
| + | |||
| + | Let us continue with the correct case. | ||
| + | As the argument is the filename of the data file, the file extension is removed to obtain the name of the file. This name will be used later. | ||
| + | |||
| + | The status variables are tuples that contain the process id of the sub-process and its exit code. | ||
| + | If the exit code is zero, the process ended normally. Errors will result in exit codes different to zero. | ||
| + | |||
| + | |||
| + | Then all the necessary tasks are done. The following task is run if the exit code of the preceding process equals zero. | ||
| + | |||
| + | |||
| + | The order and the tasks of the called scripts and programs | ||
| + | |||
| + | *'''./process.py''' Read the data file and create the TUI script. The TUI script is a python script that tells Salome what to do. | ||
| + | |||
| + | *'''source /SALOME-ROOT/salome_5.1.2/KERNEL_5.1.2/salome.sh''' This script is necessary for Salome to start up. This is stated in the readme file of this Salome version. The command '''source''' runs the command in the same shell this script is executed. So environment variables set by salome.sh are visible to runSalome. | ||
| + | |||
| + | *'''/SALOME-ROOT/salome_5.1.2/KERNEL_5.1.2/bin/salome/runSalome -t -u ./outTUI.py''' Run a TUI script in Salome in terminal mode. | ||
| + | |||
| + | *'''./CreateJobScript.py''' Create a FE study. This script was taken from the helpers directory of CAELinux. | ||
| + | |||
| + | *'''./CreateExportScript.py''' This script creates a *.export file. This file tells CodeAster what to do. | ||
| + | |||
| + | *'''/opt/SALOME-MECA-2009.1-GPL/aster/outils/as_run --run profile''' Run the FE study described by the profile. The profile is the *.export file. | ||
| + | |||
| + | *'''cat resuFile''' Display the *.resu file. Here are our results stored. | ||
| + | |||
| + | |||
| + | === Some basic post processing tasks === | ||
| + | |||
| + | I made this script using the examples from Salome. Importing this into Salome will import the *res.med file and create plots of the Von-Mises-Stresses for each load case. | ||
| + | |||
| + | |||
| + | import salome | ||
| + | import visu_gui | ||
| + | import SALOMEDS | ||
| + | import VISU | ||
| + | |||
| + | |||
| + | #%====================Stage1: Creating a new study====================% | ||
| + | |||
| + | print "**** Stage1: Creating a new study" | ||
| + | |||
| + | print "Creating a new study..................", | ||
| + | myVisu = visu_gui.myVisu | ||
| + | myVisu.SetCurrentStudy(salome.myStudy) | ||
| + | myViewManager = myVisu.GetViewManager() | ||
| + | if myViewManager is None : print "Error" | ||
| + | else : print "OK" | ||
| + | |||
| + | #%====================Stage2: Importing MED file====================% | ||
| + | |||
| + | print "**** Stage2: Importing MED file" | ||
| + | |||
| + | print 'Import "*res.med"...............', | ||
| + | medFile = "/working_root/tool01/tool01res.med" | ||
| + | myResult = myVisu.ImportFile(medFile) | ||
| + | if myResult is None : print "Error" | ||
| + | else : print "OK" | ||
| + | |||
| + | print 'Creating new View3D...................', | ||
| + | myView = myViewManager.Create3DView() | ||
| + | if myView is None : print "Error" | ||
| + | else : print "OK" | ||
| + | |||
| + | myMeshName = 'MeshLin' | ||
| + | myCellEntity = VISU.CELL | ||
| + | myNodeEntity = VISU.NODE | ||
| + | |||
| + | #%====================Stage3: Displaying scalar map====================% | ||
| + | |||
| + | print "**** Stage3: Displaying sclar map" | ||
| + | |||
| + | print "Creating Scalar Map 1.......", | ||
| + | scalarmap1 = myVisu.ScalarMapOnField(myResult,myMeshName,myNodeEntity,'Sol1E___EQUI_NOEU_SIGM__________',1); | ||
| + | if scalarmap1 is None : print "Error" | ||
| + | else : print "OK" | ||
| + | scalarmap1.SetSize(0.15, 0.8) | ||
| + | scalarmap1.SetScalarMode(1) | ||
| + | scalarmap1.SetRange(0, 283.40904) | ||
| + | myView.DisplayOnly(scalarmap1) | ||
| + | |||
| + | myView.SetView((VISU.View3D.TOP)) | ||
| + | salome.sg.updateObjBrowser(1) | ||
| + | |||
| + | |||
| + | print "Saving Scalar Map1........." | ||
| + | savepicture = myView.SavePicture("/working_root/tool01/picture01.png") | ||
| + | if savepicture is None : print "Error" | ||
| + | else : print "OK" | ||
| + | |||
| + | |||
| + | print "Creating Scalar Map 2.......", | ||
| + | scalarmap2 = myVisu.ScalarMapOnField(myResult,myMeshName,myNodeEntity,'Sol2E___EQUI_NOEU_SIGM__________',1); | ||
| + | if scalarmap2 is None : print "Error" | ||
| + | else : print "OK" | ||
| + | scalarmap2.SetSize(0.15, 0.8) | ||
| + | scalarmap2.SetScalarMode(1) | ||
| + | scalarmap2.SetRange(0, 149.23416) | ||
| + | myView.DisplayOnly(scalarmap2) | ||
| + | |||
| + | myView.SetView((VISU.View3D.TOP)) | ||
| + | salome.sg.updateObjBrowser(1) | ||
| + | |||
| + | |||
| + | print "Saving Scalar Map2........." | ||
| + | savepicture = myView.SavePicture("/working_root/tool01/picture02.png") | ||
| + | if savepicture is None : print "Error" | ||
| + | else : print "OK" | ||
| + | |||
| + | |||
| + | #%====================Stage4: Saving study====================% | ||
| + | |||
| + | print "Saving Study.............." | ||
| + | savestudy = visu_gui.myStudyManager.SaveAs("/work/tool01/tool01_post.hdf",salome.myStudy,False) | ||
| + | if savestudy is None: | ||
| + | print "Error" | ||
| + | else: | ||
| + | print "OK" | ||
to be continued ... | to be continued ... | ||
Latest revision as of 12:52, 18 November 2010
Contents
Doing an automated study with Salome and CodeAster
This example may seem a little artificial. However this was part of my work this summer for a research partner of my university. I am not allowed to place the original problem here because conflicts with the companies secrets would be inevitable. So I have to construct a new problem around the expertise I gained from my task. Even if you do not have a similar task you may learn something.
Preface
If there is the need to analyze lots of topological likewise geometries then automation can make life easier. This paragraph describes automatable analysis of a 2D part section. Given the case the geometry comes as list of vertices in a known order we can read this list with a script and create a TUI script.
The input data
The input data for the analysis is stored in a text file containing information about the geometry and the load. The geometry, a 2D part section, is given by a list of vertices. The point where the concentrated load acts on the structure is also given. Distributed load in 3D results in concentrated load in the 2D section. The text file may come from a CAD program. At least in my work this was the case.
# data for part sect01 name = 'sect01' case1.fx = 120.0; case1.fy = 21.5; case2.fx = -54.0; case2.fy = 97.2; vrtcs = [ 12.0 100.0 ... ... ... ... ];
Processing the input data
We process the data file with a python script. This script generated the TUI script which will be processed by Salome. We could also directly import the geometry in the TUI script via exchange file format but then we would not know where to apply the load and where to constrain the degrees of freedom. At least I would not know.
Running the TUI script
Salome will be started in terminal mode and the TUI script will be directly imported by command line option. As this whole analysis is made by one python script, only the parts concerning Salome will be shown here. The whole script will be listed somewhere below.
p1 = Popen("source /home/gerhard/salome_5.1.2/KERNEL_5.1.2/salome.sh", shell=True)
sts1 = os.waitpid(p1.pid, 0)
p2 = Popen("/home/gerhard/salome_5.1.2/KERNEL_5.1.2/bin/salome/runSalome -t -u ./outTUI.py", shell=True)
sts2 = os.waitpid(p2.pid, 0)
Popen() executes the given string as a sub-process. waitpid() ensures the sub-process is finished before the calling process continues execution.
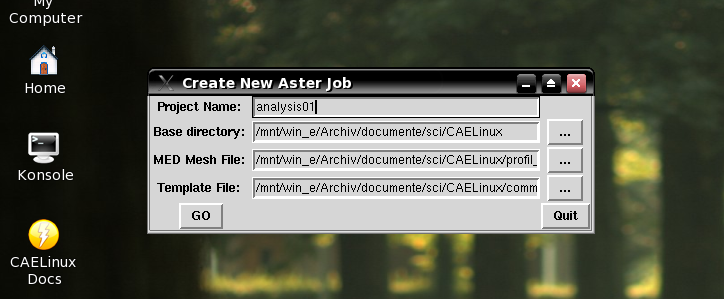
Creating the FE study
This part was taken from CAELinux. It is the core of the CreateJob.py script that uses a GUI for the major input. Here the original script is shown in action.
I removed the things related to the GUI. Crude work I have to admit. This script receives the filename of the data file as argument.
#!/usr/bin/env python # Create a new FE analysis job from a template # J.Cugnoni, CAELinux.com, 2005 # edited by G. Holzinger, JKU Linz, 2009
import os import os.path import sys
fileName = sys.argv[1]
name = fileName[:fileName.rfind(".")]
templateASTK=""" etude,fich,3,FR F opt_val,rep_dex _VIDE etude,fich,3,UL 8 etude,fich,2,type mess etude,fich,4,donnee 0 option,nbmaxnook 1 etude,fich,2,resultat 1 nom_fich_export _VIDE option,rep_dex 0 etude,fich,4,compress 0 etude oui debug 0 opt_val,cpresok RESNOOK forlib_delete non etude,fich,4,serv Local etude,fich,3,donnee 0 serv_fich_export -1 surcharge,nbfic 0 etude,fich,6,FR F path_etude [__prjdir__] option,rep_outils 0 etude,fich,6,UL 80 etude,fich,3,resultat 1 option,cpresok 1 etude,fich,5,compress 0 etude,fich,1,FR F etude,fich,1,serv Local consult_supprimer non memoire 128 etude,fich,1,UL 20 onglet_actif etude asquit non etude,fich,2,donnee 0 option,classe 1 etude,fich,4,type erre consult_a_corriger non ident non etude,fich,4,resultat 1 etude,fich,6,compress 0 etude,fich,1,donnee 1 rex non etude,fich,1,type libr etude,fich,1,nom ./[__prjname__]mesh.med suivi_interactif 1 path_sources _VIDE pre_eda non etude,fich,4,FR F forlib_create non etude,fich,4,UL 9 etude,fich,3,nom ./[__prjname__].resu serv_tests -1 etude,fich,6,serv Local etude,fich,5,nom ./[__prjname__].base make_etude run option,depart 1 nom_profil [__astkfile__] args _VIDE etude,fich,5,resultat 1 etude,fich,0,donnee 1 opt_val,mem_aster _VIDE etude,fich,3,serv Local etude,fich,0,compress 0 option,mem_aster 1 asdeno non serv_surcharge -1 serv_sources -1 surcharge non etude,fich,6,type rmed etude,fich,0,serv Local M_1 oui serv_etude -1 M_2 non etude,fich,6,resultat 1 etude,fich,2,FR F opt_val,rep_outils _VIDE M_3 non etude,fich,2,UL 6 path_surcharge _VIDE consult non M_4 non asno non etude,fich,3,type resu etude,fich,1,compress 0 opt_val,ncpus 1 emis_sans non serveur localhost opt_val,classe _VIDE opt_val,dbgjeveux _VIDE option,ncpus 1 etude,fich,0,type comm opt_val,facmtps 1 etude,fich,0,resultat 0 opt_val,rep_mat _VIDE special _VIDE tests,nbfic 0 temps 240 option,dbgjeveux 0 etude,fich,5,FR R etude,fich,5,serv Local asrest non batch 0 etude,fich,5,UL 0 etude,fich,2,compress 0 option,facmtps 1 serv_profil -1 etude,fich,6,donnee 0 etude,fich,0,FR F option,rep_mat 0 etude,fich,0,UL 1 etude,fich,2,serv Local etude,fich,0,nom ./[__prjname__].comm asverif non etude,fich,2,nom ./[__prjname__].mess opt_val,depart _VIDE etude,fich,1,resultat 0 sources,nbfic 0 etude,fich,5,donnee 0 etude,fich,4,nom ./[__prjname__].erre tests non noeud localhost etude,fich,5,type base etude,fich,3,compress 0 emis_prof non etude,fich,6,nom ./[__prjname__]res.med etude,nbfic 7 agla non opt_val,nbmaxnook 5 version STA8.1 path_tests _VIDE nom_profil %s path_etude %s etude,fich,0,nom ./%s.comm etude,fich,1,nom ./%smesh.med etude,fich,2,nom ./%s.mess etude,fich,3,nom ./%s.resu etude,fich,4,nom ./%s.erre etude,fich,5,nom ./%s.base etude,fich,6,nom ./%sres.med """
def validate():
bdir=vars["basedirname"].strip()
prjname=vars["prjname"].strip()
meshname=vars["meshname"].strip()
tplname=vars["tplname"].strip()
prjdir=os.path.join(bdir,prjname)
if os.path.exists(prjdir):
print "project name already exists in base directory"
else:
# project directory
os.makedirs(prjdir)
# file names
commfile=os.path.join(prjdir,prjname + ".comm")
messfile=os.path.join(prjdir,prjname + ".mess")
errefile=os.path.join(prjdir,prjname + ".erre")
resufile=os.path.join(prjdir,prjname + ".resu")
mmedfile=os.path.join(prjdir,prjname + "mesh.med")
rmedfile=os.path.join(prjdir,prjname + "res.med")
basefile=os.path.join(prjdir,prjname + ".base")
astkfile=os.path.join(prjdir,prjname + ".astk")
# copy files
from shutil import copyfile
copyfile(meshname,mmedfile)
copyfile(tplname,commfile)
# create ASTK profile
fd=open(astkfile,"w")
fd.write(templateASTK % ((astkfile,prjdir,) + (prjname,)*7))
fd.close()
# show message and exit
print "New FE analysis project created successfully."
return 0
vars={"basedirname": os.getcwd() + "/",
"prjname": name,
"meshname": "mesh_" + name + ".med",
"tplname": "command.comm"}
validate()
Create the *.export file
For this task there is also a script. This script receives the name of the study as argument. To adjust this file for your needs compare the output of this script with a *.export file from a running CodeAster study created by ASTK. Adjust everything you need to obtain equal *.export files.
#!/usr/bin/env python # # Create an .export-file for CodeAster # G. Holzinger, JKU Linz, 2009
import sys import os
user = os.getlogin() machine = os.uname()[1] workingRoot = os.getcwd()
# read argument name = sys.argv[1]
# open the file
f = open("./" + name + "/" + name + ".export",'w')
f.write("P profastk " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + ".astk\n")
f.write("P serveur localhost\n")
f.write("P noeud " + machine + "\n")
f.write("P username " + user + "\n")
f.write("P mclient " + machine + "\n")
f.write("P uclient " + user + "\n")
f.write("P version STA9.4\n")
f.write("P lang en\n")
f.write("P debug nodebug\n")
f.write("P mode interactif\n")
f.write("P ncpus 1\n")
f.write("P mpi_nbcpu 1\n")
f.write("P mpi_nbnoeud 1\n")
f.write("P classe \n")
f.write("P depart \n")
f.write("P distrib \n")
f.write("P timeout \n")
f.write("P exectool \n")
f.write("P nomjob " + name + "\n")
f.write("P origine ASTK 1.7.1\n")
f.write("P display " + machine + ":0.0\n")
f.write("A args \n")
f.write("A memjeveux 32.0\n")
f.write("P mem_aster 100.0\n")
f.write("A tpmax 240\n")
f.write("P memjob 131072\n")
f.write("P tpsjob 4\n")
f.write("P nbmaxnook 5\n")
f.write("P cpresok RESNOOK\n")
f.write("P facmtps 1\n")
f.write("P corefilesize unlimited\n")
f.write("F comm " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + ".comm D 1\n")
f.write("F libr " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + "mesh.med D 20\n")
f.write("F mess " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + ".mess R 6\n")
f.write("F resu " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + ".resu R 8\n")
f.write("F erre " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + ".erre R 9\n")
f.write("R base " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + ".base R 0\n")
f.write("F rmed " + user + "@" + machine + ":" + workingRoot + ("/" + name)*2 + "res.med R 80\n")
f.write("P consbtc oui\n")
f.write("P soumbtc oui\n")
f.write("P actions make_etude")
# close the file f.close()
Run the FE study with Code Aster
Here are only the few lines related to CodeAser shown, the whole script is listed further below.
exportFullPath = os.getcwd() + ("/" + name)*2 + ".export"
p5 = Popen("/opt/SALOME-MECA-2009.1-GPL/aster/outils/as_run --run " + exportFullPath, shell=True)
sts5 = os.waitpid(p5.pid, 0)
Doing the study
Here is where all the magic happens. Don't get confused with all the boxes, this source code belongs to one file. However these boxes subdividing the code into blocks may improve readability.
#!/usr/bin/env python
#
# Perform a FE analysis using Salome and CodeAster
# G. Holzinger, JKU Linz, 2009
#
# The text file containing the geometry data must reside in the same directory as this file.
# Provide the filename as argument!
# If the filename contains dots ('.') then the file must have a file extension (i.e. sect01.txt or sect01.v02.txt)
# Files without file extension are also allowed (i.e. sect01)
# Not allowed are files with filenames containing dots but without file extension (i.e. sect01.v02) for
# everything left of the rightmost dot excluding this dot is used as projectname and is used to build
# the file paths too. This would not work in this case!
import os import sys from subprocess import *
def usage(): print "\tusage: ./start.py FILENAME" print "" print "\texample: ./start.py sect01.txt"
# retrieve number of arguments nargs = len(sys.argv)
# no argument was passed # nargs == 1 because the filename of the file itself is automatically passed as argument if nargs == 1: print "no arguments passed!" usage()
# one argument was passed
if nargs == 2:
argument = sys.argv[1]
# remove file extension
index = sys.argv[1].rfind(".")
if index == -1:
name = sys.argv[1]
else:
name = sys.argv[1][0:index]
print name
# initialize status variables
sts0 = (0, 1)
sts1 = (0, 1)
sts2 = (0, 1)
sts3 = (0, 1)
sts4 = (0, 1)
sts5 = (0, 1)
# create TUI script for salome
print "calling: ./process.py " + argument
p0 = Popen("./process.py " + argument, shell=True)
sts0 = os.waitpid(p0.pid, 0)
if sts0[1] == 0:
# create mesh
p1 = Popen("source /SALOME-ROOT/salome_5.1.2/KERNEL_5.1.2/salome.sh; /SALOME-ROOT/salome_5.1.2/KERNEL_5.1.2/bin/salome/runSalome -t -u ./outTUI.py", shell=True)
sts1 = os.waitpid(p1.pid, 0)
if sts1[1] == 0:
# create finite element study
p3 = Popen("./CreateJobScript.py " + argument, shell=True)
sts3 = os.waitpid(p3.pid, 0)
if sts3[1] == 0:
# create the export file
p4 = Popen("./CreateExportScript.py " + name, shell=True)
sts4 = os.waitpid(p4.pid, 0)
# run the study
exportFullPath = os.getcwd() + ("/" + name)*2 + ".export"
resuFullPath = os.getcwd() + ("/" + name)*2 + ".resu"
p5 = Popen("/opt/SALOME-MECA-2009.1-GPL/aster/outils/as_run --run " + exportFullPath, shell=True)
sts5 = os.waitpid(p5.pid, 0)
if sts5[1] == 0:
# show results
p6 = Popen("cat " + resuFullPath, shell=True)
sts6 = os.waitpid(p6.pid, 0)
# end if
# end if
# end if
# end if
# print status report
print "return values of called scripts and programs\n\n"
print "process.py: " + str(sts0)
if sts0[1] == 0:
print "\tOk\n"
else:
print "\tFailed\n"
print "salome.sh: " + str(sts1)
if sts1[1] == 0:
print "\tOk\n"
else:
print "\tFailed\n"
print "runSalome: " + str(sts2)
if sts2[1] == 0:
print "\tOk\n"
else:
print "\tFailed\n"
print "CreateJobScript.py: " + str(sts3)
if sts3[1] == 0:
print "\tOk\n"
else:
print "\tFailed\n"
print "CreateExportScript.py: " + str(sts4)
if sts4[1] == 0:
print "\tOk\n"
else:
print "\tFailed\n"
print "as_run: " + str(sts5)
if sts5[1] == 0:
print "\tOk\n"
else:
print "\tFailed\n"
if nargs > 2: print "too many arguments passed!" usage()
The first Block is made up of comments. The most important thing here is the first line. It tells the linux shell what to do with this script.
In the second Block several python modules are imported.
The third block defines a functions that prints the usage information. Rudimentay though.
In the fourth block the number of arguments is determined.
The next three Blocks are dedicated to three possible cases. No arguments were passed, one - which is the correct number - or too many. If the wrong number of arguments passed is wrong, then error messages will be printed.
Let us continue with the correct case. As the argument is the filename of the data file, the file extension is removed to obtain the name of the file. This name will be used later.
The status variables are tuples that contain the process id of the sub-process and its exit code. If the exit code is zero, the process ended normally. Errors will result in exit codes different to zero.
Then all the necessary tasks are done. The following task is run if the exit code of the preceding process equals zero.
The order and the tasks of the called scripts and programs
- ./process.py Read the data file and create the TUI script. The TUI script is a python script that tells Salome what to do.
- source /SALOME-ROOT/salome_5.1.2/KERNEL_5.1.2/salome.sh This script is necessary for Salome to start up. This is stated in the readme file of this Salome version. The command source runs the command in the same shell this script is executed. So environment variables set by salome.sh are visible to runSalome.
- /SALOME-ROOT/salome_5.1.2/KERNEL_5.1.2/bin/salome/runSalome -t -u ./outTUI.py Run a TUI script in Salome in terminal mode.
- ./CreateJobScript.py Create a FE study. This script was taken from the helpers directory of CAELinux.
- ./CreateExportScript.py This script creates a *.export file. This file tells CodeAster what to do.
- /opt/SALOME-MECA-2009.1-GPL/aster/outils/as_run --run profile Run the FE study described by the profile. The profile is the *.export file.
- cat resuFile Display the *.resu file. Here are our results stored.
Some basic post processing tasks
I made this script using the examples from Salome. Importing this into Salome will import the *res.med file and create plots of the Von-Mises-Stresses for each load case.
import salome import visu_gui import SALOMEDS import VISU
#%====================Stage1: Creating a new study====================%
print "**** Stage1: Creating a new study"
print "Creating a new study..................", myVisu = visu_gui.myVisu myVisu.SetCurrentStudy(salome.myStudy) myViewManager = myVisu.GetViewManager() if myViewManager is None : print "Error" else : print "OK"
#%====================Stage2: Importing MED file====================%
print "**** Stage2: Importing MED file"
print 'Import "*res.med"...............', medFile = "/working_root/tool01/tool01res.med" myResult = myVisu.ImportFile(medFile) if myResult is None : print "Error" else : print "OK"
print 'Creating new View3D...................', myView = myViewManager.Create3DView() if myView is None : print "Error" else : print "OK"
myMeshName = 'MeshLin' myCellEntity = VISU.CELL myNodeEntity = VISU.NODE
#%====================Stage3: Displaying scalar map====================%
print "**** Stage3: Displaying sclar map"
print "Creating Scalar Map 1.......", scalarmap1 = myVisu.ScalarMapOnField(myResult,myMeshName,myNodeEntity,'Sol1E___EQUI_NOEU_SIGM__________',1); if scalarmap1 is None : print "Error" else : print "OK" scalarmap1.SetSize(0.15, 0.8) scalarmap1.SetScalarMode(1) scalarmap1.SetRange(0, 283.40904) myView.DisplayOnly(scalarmap1)
myView.SetView((VISU.View3D.TOP)) salome.sg.updateObjBrowser(1)
print "Saving Scalar Map1........."
savepicture = myView.SavePicture("/working_root/tool01/picture01.png")
if savepicture is None : print "Error"
else : print "OK"
print "Creating Scalar Map 2.......", scalarmap2 = myVisu.ScalarMapOnField(myResult,myMeshName,myNodeEntity,'Sol2E___EQUI_NOEU_SIGM__________',1); if scalarmap2 is None : print "Error" else : print "OK" scalarmap2.SetSize(0.15, 0.8) scalarmap2.SetScalarMode(1) scalarmap2.SetRange(0, 149.23416) myView.DisplayOnly(scalarmap2)
myView.SetView((VISU.View3D.TOP)) salome.sg.updateObjBrowser(1)
print "Saving Scalar Map2........."
savepicture = myView.SavePicture("/working_root/tool01/picture02.png")
if savepicture is None : print "Error"
else : print "OK"
#%====================Stage4: Saving study====================%
print "Saving Study.............."
savestudy = visu_gui.myStudyManager.SaveAs("/work/tool01/tool01_post.hdf",salome.myStudy,False)
if savestudy is None:
print "Error"
else:
print "OK"
to be continued ...